Success Stories : Substrate prediction for enzyme Tm0936
A pressing challenge in biology is predicting the function of the proteins the genes encode. It would be useful to be able to probe for such function directly, based on protein structures. In an effort to do so, Johannes Hermann in the Shoichet Lab attempted to predict substrates of the enzyme Tm0936 from T. maritima. The x-ray structure of this enzyme had been determined as part of a structural genomics effort (PDB codes 1p1m and 1j6p), and it can be assigned to the Amidohydrolase Superfamily (AHS) by fold classification and by the identity of certain active site groups; these metaloenzymes are united by the attack of a nucleophilic hydroxide or water on an electrophilic center-beyond this, their mechanisms are quite diverse. Consequently, the substrates for Tm0936 were anything but clear. By sequence similarity, Tm0936 most resembles the large chlorohydrolase and cytosine deaminase subgroup, which is often used to annotate amidohydrolases of unknown function. However, Raushel's group had tested 14 cytosine derivatives as Tm0936 substrates but observed no turnover.
Site & Ligand Innovations
In an effort to find the true substrate, Johannes therefore docked a database of high-energy intermediates into the structure of Tm0936, sampling thousands of configurations and conformations of each molecule. Each of these was scored by electrostatic and van der Waals complementarity, corrected for ligand desolvation energy, and ranked accordingly. There were two important innovations here: first, the decision to restrict ourselves to the KEGG metabolites database of about 10,000 possible molecules, and second to dock these in their high-energy intermediate forms, and not in their ground state structures. We reasoned that we were more likely to find substrates among primary metabolites than in any other source of compounds, thus dramatically reducing the search space. We further reasoned that the enzyme was pre-organized to recognize the excited, high-energy intermediate form of the substrate, the docking of which would inform us of the reaction being performed in addition to improving the fidelity of the modeled interactions.
| Substrate tested | high-energy intermediate | Dock Rank | Relative docking scores (kcal mol-1) | Km (µM) | kcat (s-1) | kcat/Km (M-1s-1) |
| S-adenosyl-homocysteine | 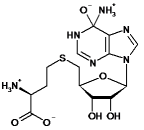 |
5 | 0 | 210±40 | 12.2±0.8 | 5.8 x 104 |
| 5-Methyl-thioadenosine | 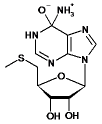 |
6 | 4.4 | 44±4 | 7.2±0.2 | 1.4 x 105 |
| Adenosine | 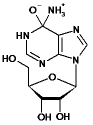 |
14 | 9.5 | 250±40 | 2.3±0.2 | 9.2 x 103 |
| adenosine-5-monophosphate | 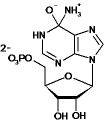 |
80 | 20.2 | nd | <10-3 | nd |
| S-adenosyl-l-methionine | 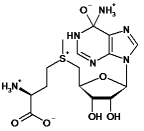 |
511 | 35.2 | nd | <10-3 | nd |